Note
Go to the end to download the full example code.
Strain Mapping#
Strain mapping in pyxem is done by fitting a DisplacementGradientMap to the data.
This can be thought of as image distortion around some central point.
from pyxem.data import simulated_strain
import hyperspy.api as hs
In this example we will create a simulated strain map using the simulated_strain() function.
This just creates a simulated diffraction pattern and applies a simple “strain” to it. In this
case using simulated data is slightly easier for demonstration purposes. If you want to use
real data the zrnb_precipitate() dataset is a good example of strain from a precipitate.
strained_signal = simulated_strain(
navigation_shape=(32, 32),
signal_shape=(512, 512),
disk_radius=20,
num_electrons=1e5,
strain_matrix=None,
lazy=True,
)
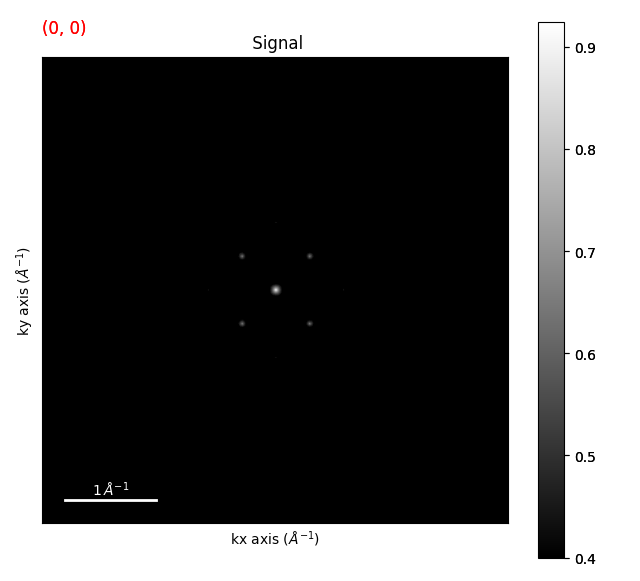
The first thing we want to do is to find peaks within the diffraction pattern. I’d recommend
using the get_diffraction_vectors() method
strained_signal.calibration.center = (
None # First set the center to be (256, 256) or the center of the signal
)
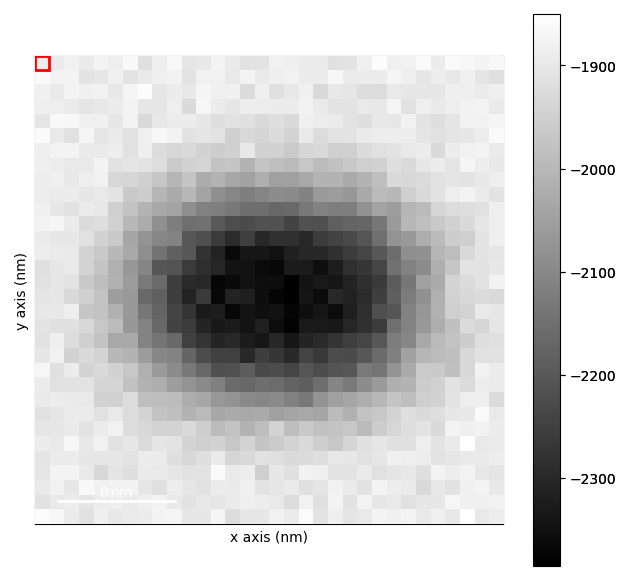
template_matched = strained_signal.template_match_disk(disk_r=20, subtract_min=False)
template_matched.plot(vmin=0.4)
0%| | 0/65 [00:00<?, ?it/s]
2%|▏ | 1/65 [00:00<00:50, 1.28it/s]
14%|█▍ | 9/65 [00:01<00:08, 6.31it/s]
26%|██▌ | 17/65 [00:02<00:06, 7.95it/s]
38%|███▊ | 25/65 [00:03<00:04, 8.73it/s]
51%|█████ | 33/65 [00:04<00:03, 9.17it/s]
63%|██████▎ | 41/65 [00:04<00:02, 9.53it/s]
75%|███████▌ | 49/65 [00:05<00:01, 9.74it/s]
88%|████████▊ | 57/65 [00:06<00:00, 9.90it/s]
100%|██████████| 65/65 [00:06<00:00, 10.09it/s]
Plotting the template matched signal and setting vmin is a good way to see what threshold you
should use for the get_diffraction_vectors() method.
diffraction_vectors = template_matched.get_diffraction_vectors(
threshold_abs=0.4, min_distance=5
)
markers = diffraction_vectors.to_markers(color="w", sizes=5, alpha=0.5)
strained_signal.plot()
strained_signal.add_marker(markers)
0%| | 0/65 [00:00<?, ?it/s]
100%|██████████| 65/65 [00:00<00:00, 1148.50it/s]
Determining the Strain#
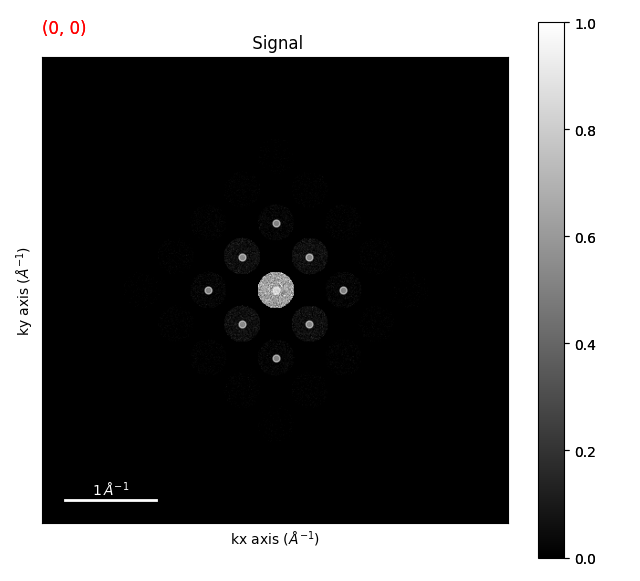
We can just use the first ring of the diffraction pattern to determine the strain. We can do this by
using the filter_magnitude() method. You can also look at the
filtering vectors example to see
how to select which vectors you want to use more generally. You can also just manually input the un-strained
vectors or use simulated/ rotated vectors as well.
first_ring_vectors = diffraction_vectors.filter_magnitude(
min_magnitude=0.1,
max_magnitude=1,
)
unstrained_vectors = first_ring_vectors.inav[0, 0]
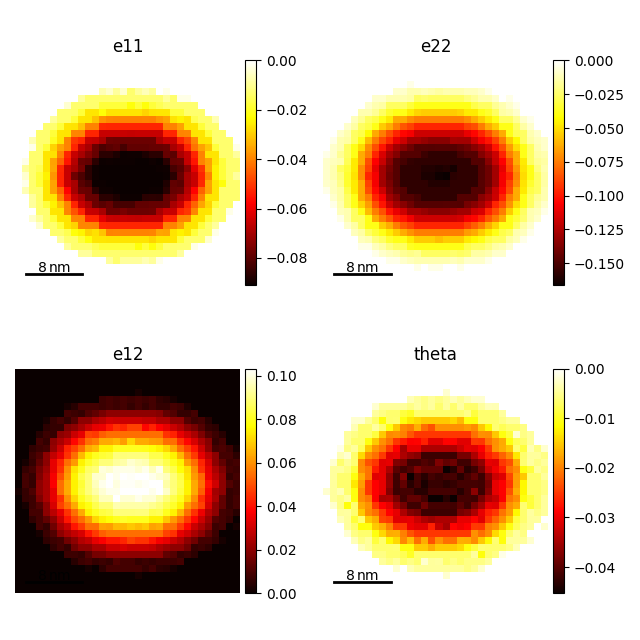
strain_maps = first_ring_vectors.get_strain_maps(
unstrained_vectors=unstrained_vectors, return_residuals=False
)
strain_maps.plot()
0%| | 0/2 [00:00<?, ?it/s]
50%|█████ | 1/2 [00:00<00:00, 1.40it/s]
100%|██████████| 2/2 [00:00<00:00, 2.80it/s]
0%| | 0/65 [00:00<?, ?it/s]
2%|▏ | 1/65 [00:01<01:11, 1.12s/it]
14%|█▍ | 9/65 [00:02<00:12, 4.54it/s]
26%|██▌ | 17/65 [00:03<00:08, 5.76it/s]
38%|███▊ | 25/65 [00:04<00:06, 6.06it/s]
51%|█████ | 33/65 [00:05<00:04, 6.48it/s]
63%|██████▎ | 41/65 [00:06<00:03, 6.64it/s]
75%|███████▌ | 49/65 [00:07<00:02, 6.85it/s]
88%|████████▊ | 57/65 [00:09<00:01, 6.99it/s]
100%|██████████| 65/65 [00:09<00:00, 7.12it/s]
0%| | 0/51 [00:00<?, ?it/s]
100%|██████████| 51/51 [00:00<00:00, 529.15it/s]
0%| | 0/51 [00:00<?, ?it/s]
100%|██████████| 51/51 [00:00<00:00, 616.18it/s]
0%| | 0/51 [00:00<?, ?it/s]
100%|██████████| 51/51 [00:00<00:00, 5497.69it/s]
0%| | 0/9 [00:00<?, ?it/s]
100%|██████████| 9/9 [00:00<00:00, 8105.81it/s]
[<Axes: title={'center': 'e11'}, xlabel='x axis (nm)', ylabel='y axis (nm)'>, <Axes: title={'center': 'e22'}, xlabel='x axis (nm)', ylabel='y axis (nm)'>, <Axes: title={'center': 'e12'}, xlabel='x axis (nm)', ylabel='y axis (nm)'>, <Axes: title={'center': 'theta'}, xlabel='x axis (nm)', ylabel='y axis (nm)'>]
Some final notes about strain mapping. In general, you want to use as many pixels as possible. 512 x 512 is a good place to start. You can do strain mapping with fewer pixels, but the results will be less accurate. Precession also helps improve the results as does having a thinner sample both of which reduce the effects of dynamical diffraction.
sphinx_gallery_thumbnail_number = 5
Total running time of the script: (0 minutes 32.235 seconds)